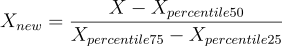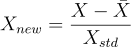Model Configurations: Anomaly Detection (Autoencoder)
When you select the Anomaly Detection (Autoencoder) modeling method Model > Configurations, the Model Configuration area of the <output> tab contains the following elements. For each output there is a separate tab.
See Model Configurations (ASCMO-DYNAMIC), for a description of the Output Properties area and the button row on the bottom of the window.
Training Labels
Assign the labels you want to train the model on. If you use multiple labels, all data associated with at least one of the labels is used.
Assign a label by double-clicking the field and typing the name. Select the dataset from the list of suggestions.
Use the x on the label or Del to remove the label.
Validation Labels
Assign the labels of the data you want to use as validation data. If you use multiple labels, all data associated with at least one of the labels is used.
Assign a label by double-clicking the field and typing the name. Select the dataset from the list of suggestions.
Use the x on the label or Del to remove the label.
|
Note |
|---|
|
If the validation labels are not assigned to any data, the model is trained without validation. A message appears in the log window. You can assign labels to data in the Manage Datasets window. |
Network Layout
Number of Layers
Enter the number of layers.
Cell Memory Size
Enter the cell memory size per node of the neural network. Typical value range is [5, 20]. The actual memory size depends on the cell type.
Last layer: The cell memory size is the number of inputs and cannot be changed.
First and third layers: The cell memory size is twice the number of inputs.
Second layer: The cell memory size is the number of inputs minus 1, called the bottleneck.
More or fewer layers can be used, but a bottleneck is always required, and the last layer always has the number of input neurons.
Note that the memory size of one cell must be less than the total number of inputs.
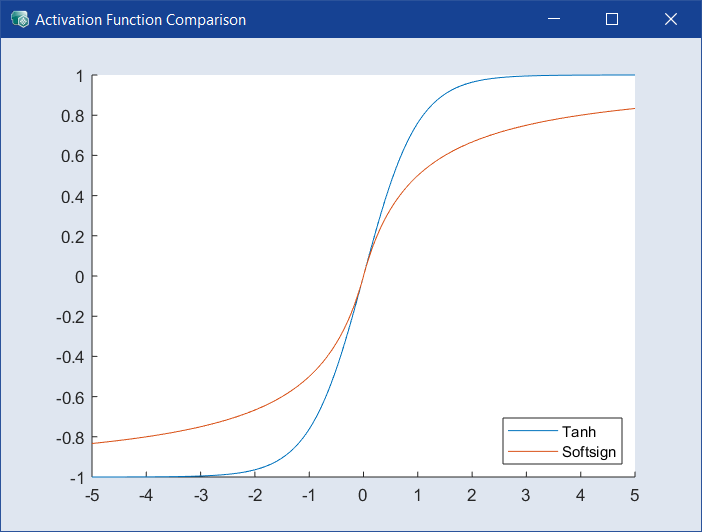
Select the activation function for all layers. Default is Tanh, a different function may be less accurate but more efficient to calculate. Click ![]() to the right of the drop-down to open the
to the right of the drop-down to open the  Activation Function Comparison window.
Activation Function Comparison window.
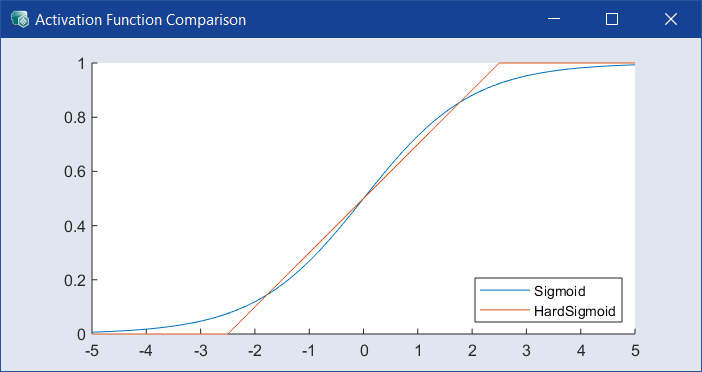
Select the recurrent activation function for all layers. Default is Sigmoid, a different function my be less accurate but more efficient to calculate. Click ![]() to the right of the drop-down to open the
to the right of the drop-down to open the  Activation Function Comparison window.
Activation Function Comparison window.
Number of Network Parameters
Dynamically shows the number of parameters used by the model training for current settings
Training Properties
Continue Training
Activate the checkbox to continue with existing model training and iterations, if possible, instead of starting a new training. You can change the training properties and continue.
Number of Multistarts
Enter the number of training repetitions with different starting values. A higher value can improve the model quality, but the model training then takes more time. The default value is 3.
Enter the number of iterations to be performed during model training. If the model performance does not improve within 10 iterations on the validation data, the training will be aborted. In deep learning this is often referred to as number of epochs.
Enter the size of sequences into which the data is segmented. The underlying optimizer then gets batches with sequences of this fixed length. The default value is 100.
If the length of the present data is about this number of steps, or less, it is recommended that you reduce the lookback length, even though an optimization with Lookback Length=100 is possible. A smaller value trains faster.
Enter a value between 0 and 0.9 as a percentage of neurons to be randomly deactivated during training. That means the weight of the neurons is set to 0 temporarily. This avoids overfitting. The entered value 0 means not activated, 0.1 means 10% of the neurons are set to 0 temporarily. The typical value range is [0, 0.2].
Enter a value between 0 and 0.9 as a percentage of recurrent state neurons to be deactivated during training. That means the weight of the neurons is set to 0 temporarily. This avoids overfitting. The entered value 0 means not activated, 0.1 means 10% of the neurons are set to 0 temporarily. The typical value range is [0, 0.2].
Plot RMSE during Training
Activate if you want the RMSE values for training data and validation data to be  displayed during model training.
displayed during model training.
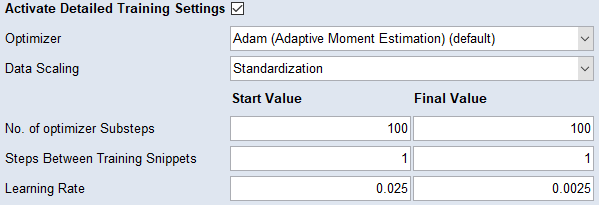
Activate Detailed Training Settings
Activate the checkbox to display the  Detailed Training Settings section.
Detailed Training Settings section.
Anomaly Percentile
Enter the percentile of reconstruction errors that are considered normal. If validation datasets are used for model selection, this value is calculated based on validation data, otherwise based on training data. The corresponding value is mapped to 0.5 in the anomaly score.
Use Signalwise Percentile Values
Activate if you want the error threshold to be calculated for each signal separately and the total score to be set to the maximum of the individual scores.
Smoothing
Enter the window size value for data points as steps for a median filter. For the prediction of an anomaly, the signals can be smoothed. This can lead to better results.
Smoothing Window Centered
Activate if you want the smoothing window to be centered around the evaluation point, i.e. future points are also taken into account. If deactivated, only past points are used for calculations in the smoothing window.
Round Score
If enabled, the score is set to 0 if it is less than the threshold, otherwise it is set to 1.
When deactivated, the score is continuously in [0,1].
Rounding Threshold
Enter an anomaly prediction threshold below which the model prediction is automatically set to 0, and 1 otherwise. The threshold is displayed in the Receiver Operating Characteristic (Model > Anomaly Detection: Receiver Operating Characteristic).
Inputs used in Model Training
This area lists all inputs of the model. You can activate/deactivate the checkbox of an input to include/exclude it in the model training.
By default, all inputs are included.
See also
Model Configurations (ASCMO-DYNAMIC)
Model Configurations: Recurrent Neural Network (RNN)
.png)
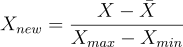 scaled to an interval of length one contained in [-1, 1].
scaled to an interval of length one contained in [-1, 1].